Welcome to VirAnnot’s documentation!¶
VirAnnot was build to ease the assembly, blast search and taxonomic annotation of metagenomic multi-sample datasets. It is used in the Virologie team of UMR1332 BFP laboratory at INRA.
It was designed to identify viruses in plants but it can be used to assemble and then annotate any sequences with the NCBI taxonomy.
NR and NT must be present localy and/or on distant servers and NCBI taxonomy is loaded in SQLITE database with a provided script.
Blast step is the most time consumming step and the use of large computer cluster is clearly an advantage. Here we used two clusters :
CURTA at Bordeaux University.
GENOTOUL at Toulouse INRA datacenter.
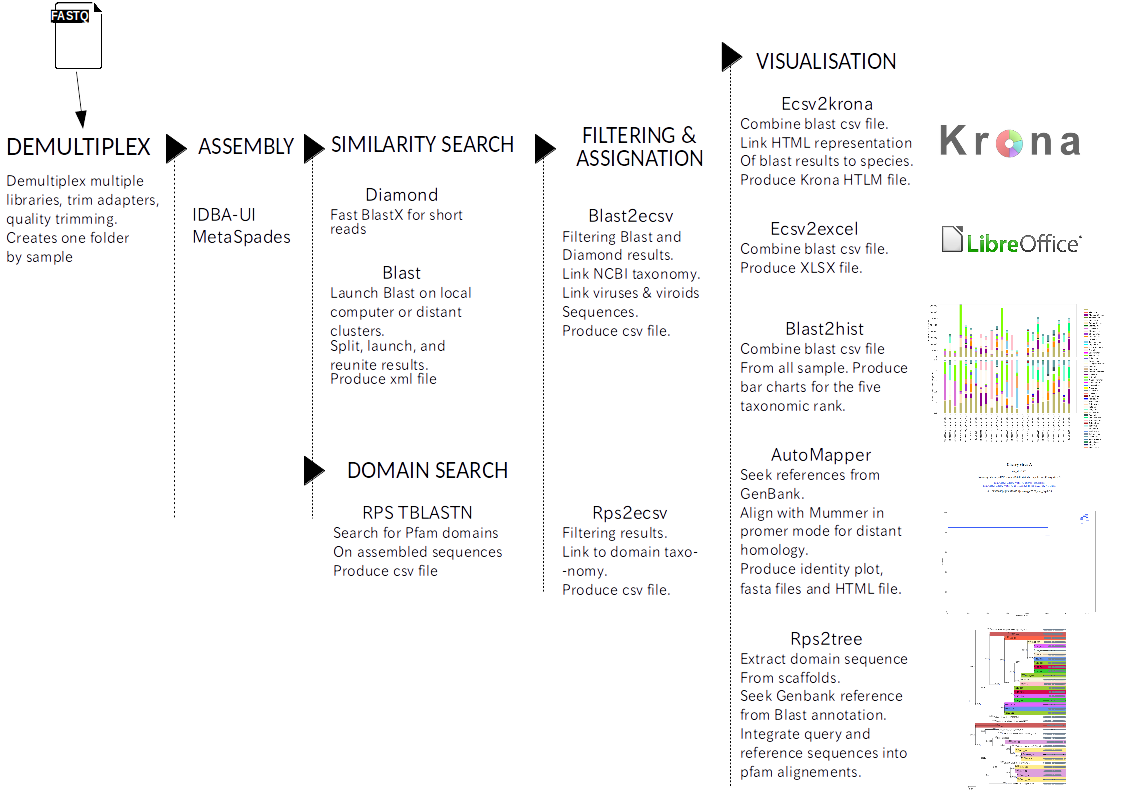
Pipeline general scheme:¶